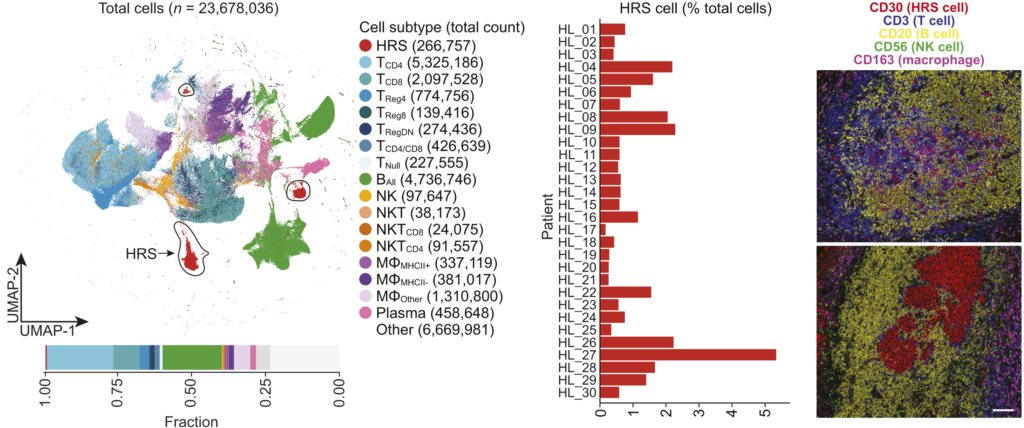
Spatial multi-omics provides granular insights into the localization of transcripts and proteins in specimens. In the current study, Maryam Pourmaleki and her colleagues investigated the heterogeneity and tumor microenvironment of classic Hodgkin lymphoma (cHL). They characterized patient-derived specimens using spatially resolved multiplexed protein imaging and transcriptomic sequencing.
Background: The study involved patient specimens derived from 36 newly diagnosed cHL tumors. The technologies employed included H&E staining, spatially resolved multiplexed protein imaging (multiplex immune fluorescence and IHC), and transcriptomic sequencing (NanoSting).
Findings: The analysis revealed several key insights.
- Hodgkin Reed-Sternberg (HRS) cells with MHC-I expression are associated with immune-inflamed neighborhoods containing CD8+ T cells and MHC-II+ macrophages.
- Spatial clustering of HRS cells was identified, particularly in syncytial variants of cHL.
- Differences in immune escape mechanisms were observed between EBV-positive and EBV-negative tumors.
- Some tumors contained regulatory T-cell–high neighborhoods with HRS cells showing increased proliferative capacity.
Reference: Pourmaleki et al, Multiplexed Spatial Profiling of Hodgkin Reed–Sternberg Cell Neighborhoods in Classic Hodgkin Lymphoma. Translational Cancer Mechanisms and Therapy, 2024. https://aacrjournals.org/clincancerres/article/30/17/3881/747268/Multiplexed-Spatial-Profiling-of-Hodgkin-Reed
