Background and Significance:
The study investigates the fibrotic response to anti-CSF-1R therapy in glioblastoma, a highly aggressive brain tumor with a poor prognosis. Despite standard treatments like surgery, chemotherapy, and radiation, glioblastomas almost always recur. This study explores an alternative strategy targeting tumor-associated macrophages and microglia through CSF-1R inhibition. Previous research showed that this approach could regress tumors and extend survival, but recurrences were still common. Understanding the mechanisms behind these recurrences is crucial for improving therapeutic outcomes.
Number of Subjects and Technology Used:
The study utilized multiple preclinical models, including transgenic mouse models and patient samples. The technology used for profiling samples included multi-omics analyses, magnetic resonance imaging (MRI), immunofluorescence (IF) imaging, single-cell RNA sequencing (scRNA-seq), and spatial transcriptomics.
Characterization of fibrotic niches around CSF-1R treated glioma tissues:
The study found that anti-CSF-1R therapy and other glioblastoma treatments triggered a fibrotic response, creating niches that promoted tumor cell survival and recurrence. These fibrotic areas were identified as pro-tumor survival niches that encapsulated surviving glioma cells, promoted dormancy, and inhibited immune surveillance.
Bioinformatics analysis was used to identify and characterize the niche regions associated with recurrence of gliomas. Hyperplexed Immunofluorescence Imaging (HIFI), a digital pathology technique (left panel) and Mass spec proteomics (right panel) was used to spatially localize protein markers associated with fibrotic niche and quantify them.
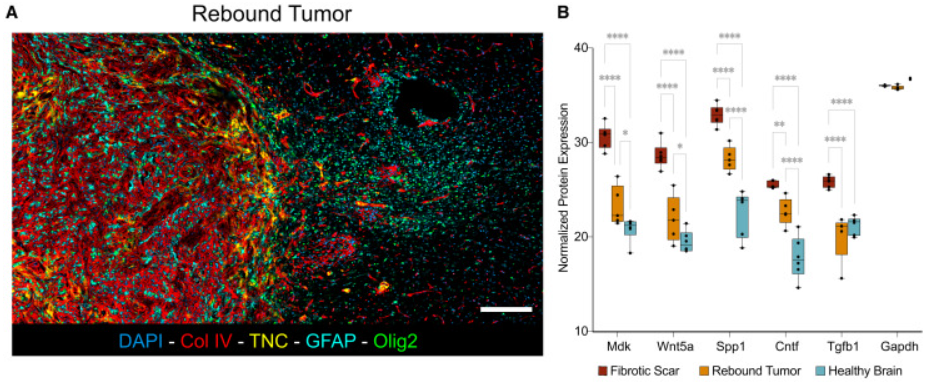
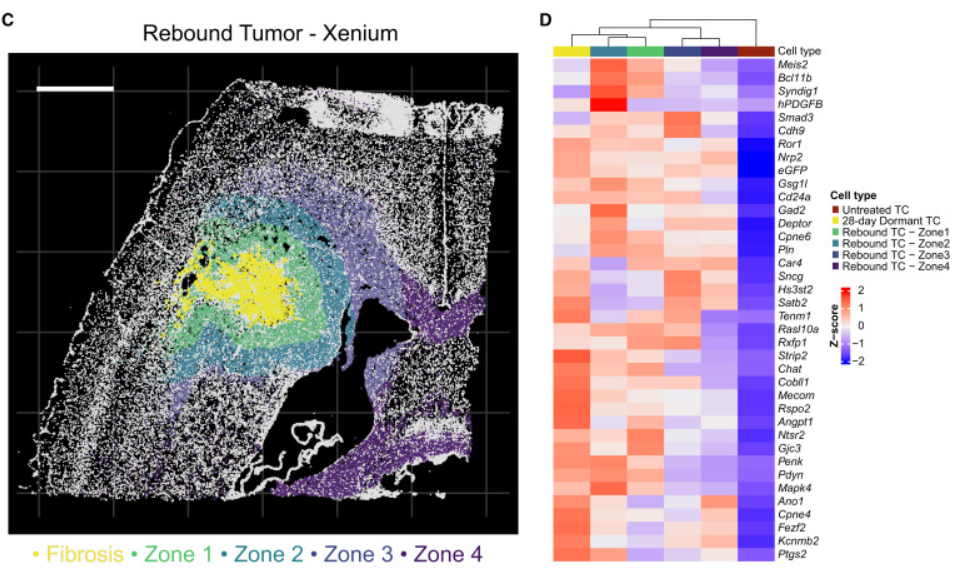
Further analysis of the tissue using Xenium scRNA-seq data identified transcriptional changes and differentially expressed genes within the different zones around the fibrotic, recurrent and normal brain tissue.
Reference: https://www.cell.com/cancer-cell/fulltext/S1535-6108(24)00311-8
